News
We thank Shakunthala Natarajan for updating latest news about the group

🌿 Kicking off Plant Bioinformatics! 💻🧬
Our first lecture in Plant Bioinformatics sets the stage for understanding how biology, big data, and computer science come together to analyze complex biological data.
We explore:
🔬 Why bioinformatics is essential in the age of massive omics datasets
🧬 Genomics, transcriptomics, proteomics & metabolomics
🧪 Sequence analysis tools like BLAST and principles of functional annotation
📊 Statistics, phylogenetics, and systems biology
💻 Programming languages (Python, R, Perl, Java) and workflow platforms like Galaxy
This lecture also covers reproducibility, version control, licensing, research data repositories, and sustainable software development — essential skills for modern life scientists. Plant science is becoming increasingly data-driven — and bioinformatics provides the toolkit to turn data into biological insight.
Cookbook for Plant Genome Sequences
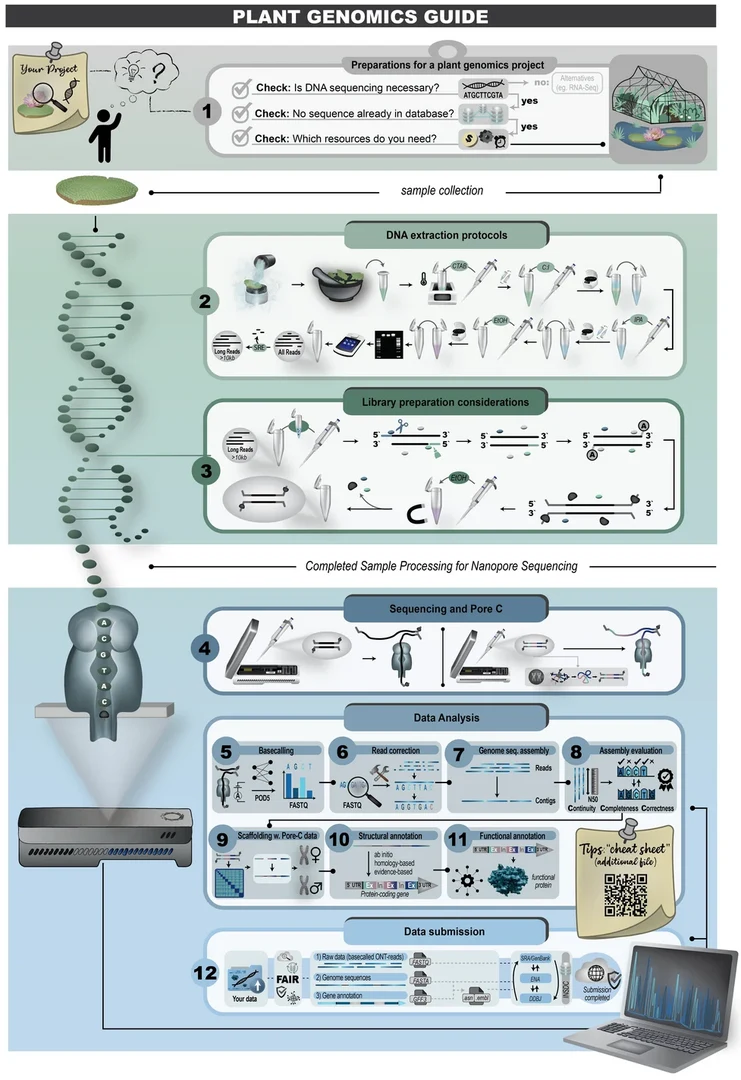
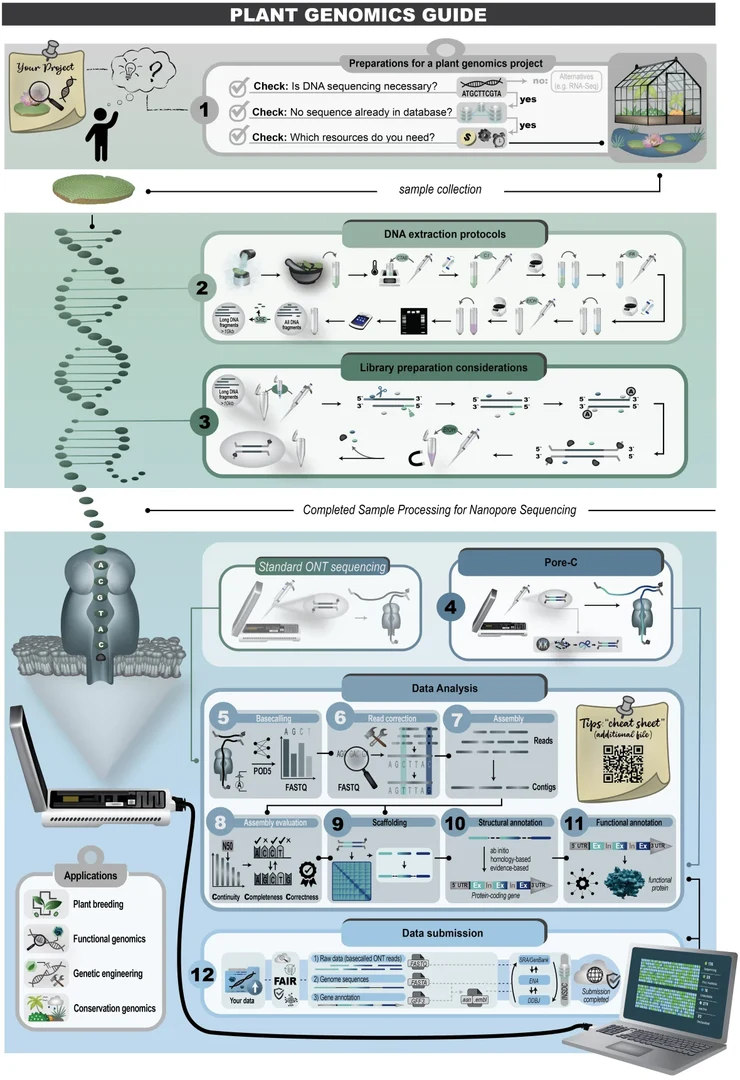
The workflow for long-read plant genomics established in our group was published in BMC Genomics to make it accessible to the whole plant science community. The steps from planning your project to finally submitting all your data to appropriate repositories are covered. The workflow is utilizing only ONT sequencing, because this is accessible to most research groups.
If you are interested how to get a fully annotated plant genome sequence, consider reading this article:
de Oliveira, J.A.V.S., Choudhary, N., Meckoni, S.N. et al. Cookbook for plant genome sequences. BMC Genomics (2026). doi: 10.1186/s12864-026-12623-z.

Celebrate the brilliance of women driving plant sciences forward at Pucker Lab on International Day of Women and Girls in Science! Here's to curiosity, innovation, and shattering glass ceilings in STEM!

Farewell for Sofi
Since Sofi is leaving us to conduct her Master thesis in Leiden, we gathered for a farewell event.

🎉 Happy New Year 2026! 🎉
We are starting the year with great news: several new publications went live over the holidays. A big thank-you to everyone involved in this work!
🔬📄 Explore the latest updates here.
Looking forward to a year full of collaboration and exciting discoveries!

Christmas party 2025
The Pucker lab is all geared up for the christmas season. We kicked off with an amazing christmas party last week. The party began with a wide spread of food and delicacies. Ranging from vegan cinnamon rolls, brownies to delicious lasagna, flavourful Indian biryani and some Brazilian and Turkish delights, the food actually represented the inclusivity and diversity in our group! We then moved on to play secret santa and everyone received a nice little surprise to take home :) Spurred by the energy from the unending food and enthusiasm of the entire team, we also had a game night involving pictionary. Thanks to the wonderful scientists in the group who are excellent organizers with great culinary skills, our first christmas party in Bonn was a day to cherish and enjoy!
Feuerzangenbowle
Last week we enjoyed a relaxed evening at the Feuerzangenbowle event hosted by the Student council biology, Bonn! The classic drink of the evening where sugarloaves were dazzling in fire was a feast to the eyes and the taste buds :) A number of other options including mulled wine and non-alcoholic punch made the event warm and welcoming for everyone. The iGEM team also joined us for the evening and we had a blast. It was a wonderful time to meet people from other groups and have fun around the festive season. The Pucker lab is looking forward to more such events in the future!
2025 was a milestone year for our group.
Our relocation to the University of Bonn opened the door to many exciting opportunities, new collaborations, and the arrival of numerous highly motivated group members.
We were proud to publish a large number of papers this year, including some of our most exciting scientific stories to date. In addition, we had the pleasure of hosting many outstanding visitors in Bonn over the past months, which greatly enriched our research environment.
We are deeply grateful to everyone who supported this transition and helped make it such a success. We look forward to building on this momentum in the years ahead.

iGEM Waffle party
Our iGEM team treated us with warm waffles last Friday! Coupled with flavourful cherry sauce and the awesome cinnamon-powdered sugar sprinkles, we had a perfect treat to our taste buds. Moreover, the iGEM team was also thoughtful enough to include vegan waffles in the menu to make it inclusive for everyone in the group. It was a perfect ending to a busy week, that was filled with the de.NBI course training sessions. Keep an eye out for our iGEM team's updates. These folks are equally adept in doing science and having fun :)
"From Gene Models to Biological Insights - Functional Annotation"
We conducted a training course about "From Gene Models to Biological Insights - Functional Annotation". Participants learned how to functionally annotate plant genes.
Slides of this course are available on GitHub: de.NBI2025.
More information about the long read genomics workflow are available in our Cookbook for Plant Genome Sequences.

"Plant Genome Sequence Assembly and Annotation" training course
We conducted a training course about "Plant Genome Sequence Assembly and Annotation". Participants learned how to assess a set of long reads, assemble a genome sequence, evaluate the assembly, and identify protein encoding genes.
Slides of this course are available on GitHub: de.NBI2025.
More information about the long read genomics workflow are available in our Cookbook for Plant Genome Sequences.

Inaugural lecture on Dies academicus
Boas gave his inaugural lecture on December 3rd. Interested members of the university are joined the lecture about "Big Data-Driven Discoveries in Specialized Plant Metabolism".

Visitor
We have a new visitor in town! We are so glad to have Leon-Dominik Dietrich from the Biochemical Ecology and Molecular Evolution group based at Christian-Albrechts-University of Kiel, as a visiting researcher. He is already having a great time in the lab with long read sequencing and off the lab with our impromptu waffle parties :)
Stay tuned for exciting updates on some interesting plant biosynthetic pathway secrets that he will be unravelling from his plant genome sequences. That being said, please note that the Pucker lab is always looking forward to host motivated students and forge fruitful collaborations. Do reach out in case you want to have fun doing some great science with a friendly group!

Christmas market visit
It's Christmas time, and of course the Pucker lab is having a great time. Our first christmas market visit on December 2nd in Bonn was filled with laughter and frolic. Along with some gluhwein we also found a perfect location for our group picture!

6th Annual Meeting of the de.NBI Industrial Forum 2025
Boas gave a talk about “Computational Approaches to Unraveling Biosynthetic Pathways in Plants” at the 6th Annual Meeting of the de.NBI Industrial Forum 2025.
Great discussions, inspiring sessions, and exciting opportunities at the intersection of data science, bioinformatics, and biotechnology.
If you see potential for collaboration or would like to explore related ideas, please feel free to reach out.

FAIR Software Award Lower Saxony 2025 for bHLH_annotator
Corinna and Boas won the FAIR Software Award Lower Saxony 2025 for the development of the bHLH_annotator. In November 2025, the award ceremony took place at TU Braunschweig.

PuckerLab at de.NBI & ELIXIR All Hands Meeting 2025
Maria and Katharina participated in the de.NBI & ELIXIR-DE All Hands Meeting 2025 that took place in Berlin from 19-21 November 2025. They presented the diverse projects in our lab in two comprehensive posters. Coupled with wonderful networking opportunities,an opportunity to know the state of art in the computational landscape relevant to biology was the highlight of the event. We thank the de.NBI facility for supporting our work and giving this wonderful chance to participate in the conference!

Visit at CZU (Prague)
Boas visited the CZU in Prague and gave a Long Read Genomics course. The Department of Genetics and Breeding hosted this teaching stay. A large number of highly motivated students attended the lectures and engaged in discussions. The teaching visit was funded by ERASMUS. Many thanks to Petr Sedlák and Vladimíra Sedláková for the excellent organization.

Today, the University of Bonn released a story about our work on anthocyanin loss in the Cucurbitaceae - just in time for halloween. Many thanks to the press office team for their amazing work. Check out the content on Instagram, BlueSky, or read the orignal work (Choudhary et al., 2025).
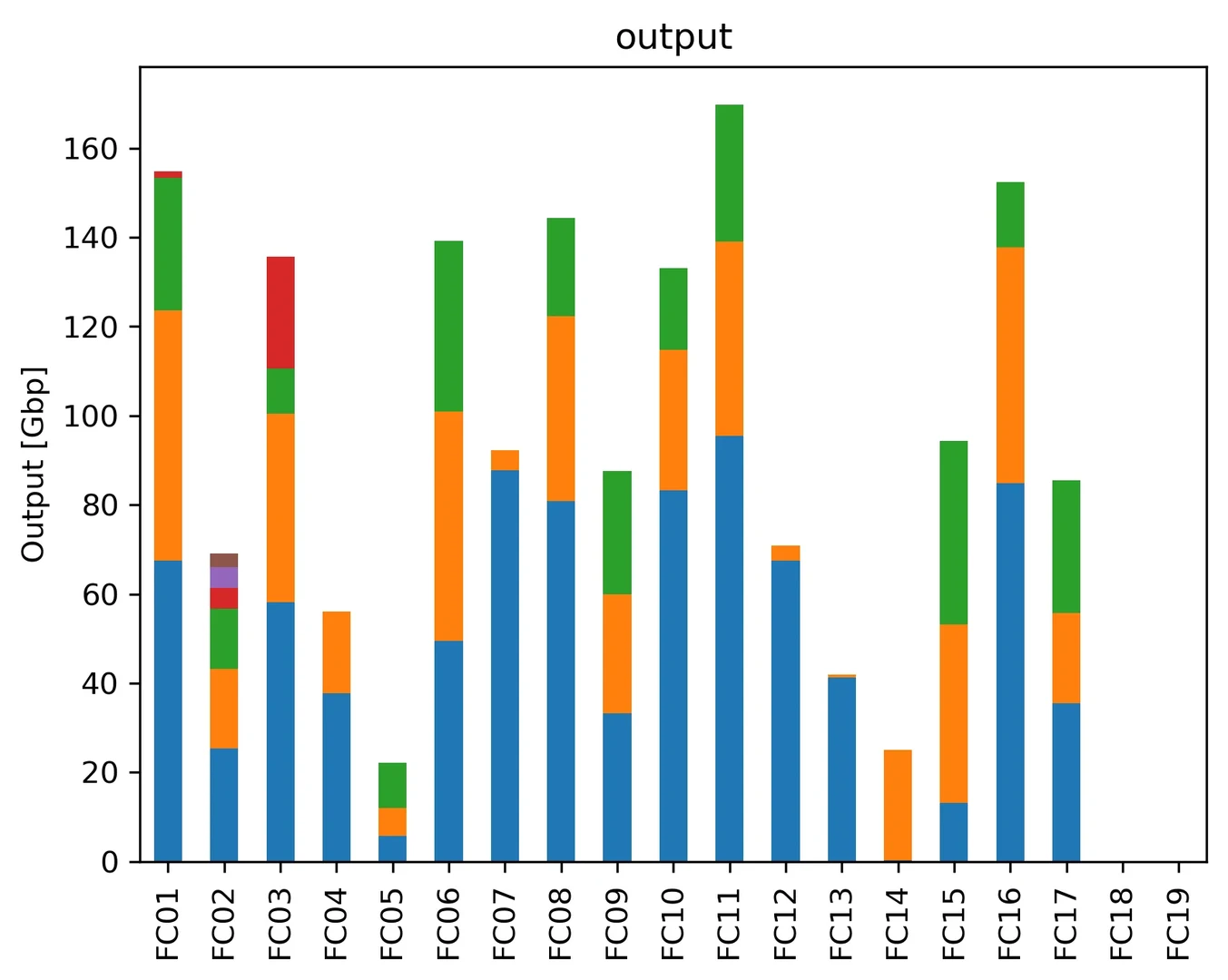
We are now performing nanopore sequencing on PromethION 2 Solo (ONT). Here, we are sharing the statistics of our output per P2 Solo flow cell. The statistics will be updated over time once the basecalling of new datasets is completed. The analysis script is available on GitHub. Details about the projects will be released soon.
DNA extraction is done with our established CTAB protocol. Long fragments are enriched via SRE. We use LSK114 for library preparation. Basecalling after the sequencing is conducted with dorado using the HAC model. The whole workflow is described in our recent review article (de Oliveira et al., 2025).

DAAD-RISE Scholarships for International Students (US, UK, Canada)
Pucker lab is excited to offer internship opportunities to students via the DAAD RISE internship program. We have had some excellent students join our group through the program and are looking forward to host curious and enthusiastic folks again this year! Our project offers are an exciting mix of bioinformatics, molecular biology and genetic engineering. If you are someone from the wet lab looking to explore computational stuff or if you are a dry lab person wanting to venture into wet lab, we have interesting research offers to pique the scientist in you. The list of projects we advertised this year are:
1) TwinFun - Exploring gene duplications in the chocolate tree (Bioinformatics)
2) Creating a living bioreactor? Utricularia research towards innovative biotechnology (Molecular biology and bioinformatics)
3) The Colors of Life: Unraveling the Genetic Palette of Plants (Bioinformatics)
4) Brewing Color in the Lab: Engineering Yeast to Synthesize Anthocyanins (Molecular biology and genetic engineering)
To know more about the projects, please visit the DAAD RISE portal.
We look forward to receiving your applications and hosting you in Bonn soon!
Supervisor spotlight
A friendly group of doctoral students from the Pucker lab will be your supervisors during the internship time. You will be working on some important parts of your supervisor's doctoral project. This is a fantastic chance for us and you to 'Train and learn with each other'. Please visit the portfolio links below to more about your prospective supervisors.
Katharina Wolff

Nanopore Sequencing in Plant Sciences
Boas is going to give a talk about our nanopore sequencing activities at a symposium in Freiburg.If you would like to read about our plant genome sequencing workflow, have a look at our cookbook for genome sequences.

Samuel is an sdw PhD scholarship holder. As an sdw scholar he gets a number of opportunities for professional and personal development. One such immensely useful event was a recent meeting held at Forschungszentrum Jülich. It was organized by the STEM section of the sdw and provided the scholars a chance to visit scientific facilities and listen to lectures on various themes. Samuel opted to visit the Exascale computing facility during the research visits organized and had a thrilling experience in the vicinity of JUPITER, Europe's first exascale supercomputer. Further he also visited another company, Microscale Bioengineering, in FZ Jülich that specializes in developing microfluidic tools for life science research. Ranging from massive supercomputing facilities to intricate micro-scale technologies, he had a scientific blast and got a fresh perspective on research and technology development. Last but not the least, he also particularly enjoyed the career talks that also focused on roles beyond traditional research positions and academia. You can find more about his visit here.



Samuel recently attended his first conference in the scenic city of roses - Hildesheim! The conference was specifically aimed at early career researchers and was organized by the German Botanical Society's (DBG's) sections - "Natural Products" and "Applied Botany". Samuel also got an opportunity to present his research through a talk titled "Blushing traps: When a carnivore goes on a nitrogen diet". Along with useful research inputs for his further work, he also gleaned some nice ideas from the talks of fellow researchers. With some good research inspirations from the conference, he is thankful to the organizers for the event, and is all set to take next strides in his research on Utricularia. Stay tuned for exciting research stories on the tiny bladderwort from him! More impressions on his conference visit can be found at: https://mstdn.science/@samnm/115151781950041399.

Harnessing Big Data to Unlock Specialized Plant Metabolism for Metabolic Engineering
Boas gave a talk at the Frauenhofer IME in Aachen. This talk covered genomics and transcriptomics presenting work on Digitalis purpurea as an example for the discovery of a gene responsible for a trait. Our work on dark pigments and the characterization of the withanolide biosynthesis were also presented. The power of coexpression analyses was demonstrated. KIPEs and CoExpPhylo served as important examples for tools to facilitate the functional annotation of plant genes.

It has been sometime since the Pucker lab shifted to Bonn and we have been having a great time here! We recently enjoyed the institute summer party at the courtyard on a perfect sunny day. Along with good food and funny banter, we had a chance to meet other colleagues here at IZMB too! Of course, we had a barbecue as well, just in case you are wondering :)